Analysis of Oral Microbiome from Fossil Human Remains Revealed the Significant Differences in Virulence Factors of Modern and Ancient Tannerella Forsythia
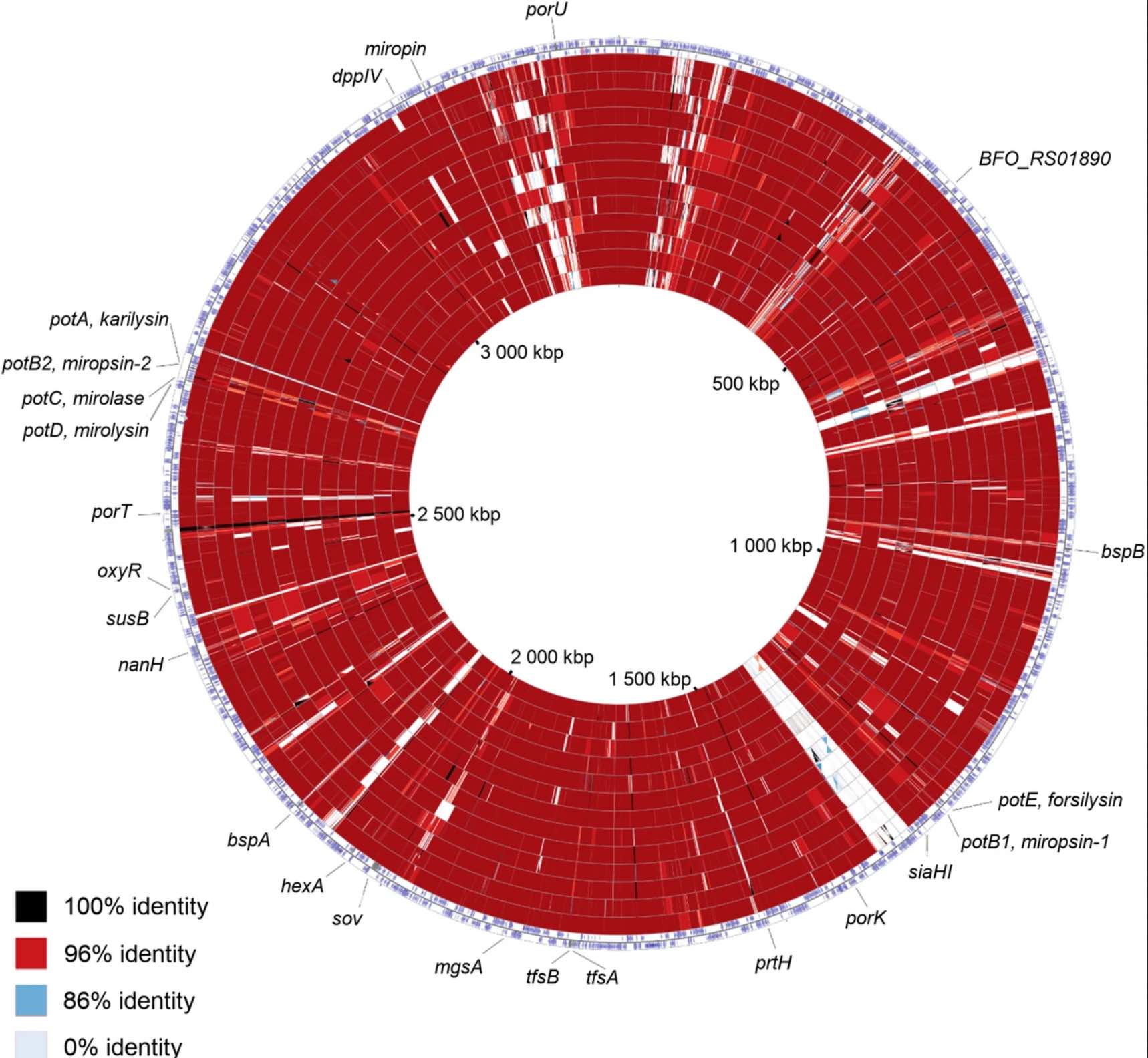 Figure: ©biomedcentral.com
Figure: ©biomedcentral.comBackground
Recent advances in the next-generation sequencing (NGS) allowed the metagenomic analyses of DNA from many different environments and sources, including thousands of years old skeletal remains. It has been shown that most of the DNA extracted from ancient samples is microbial. There are several reports demonstrating that the considerable fraction of extracted DNA belonged to the bacteria accompanying the studied individuals before their death.
Results
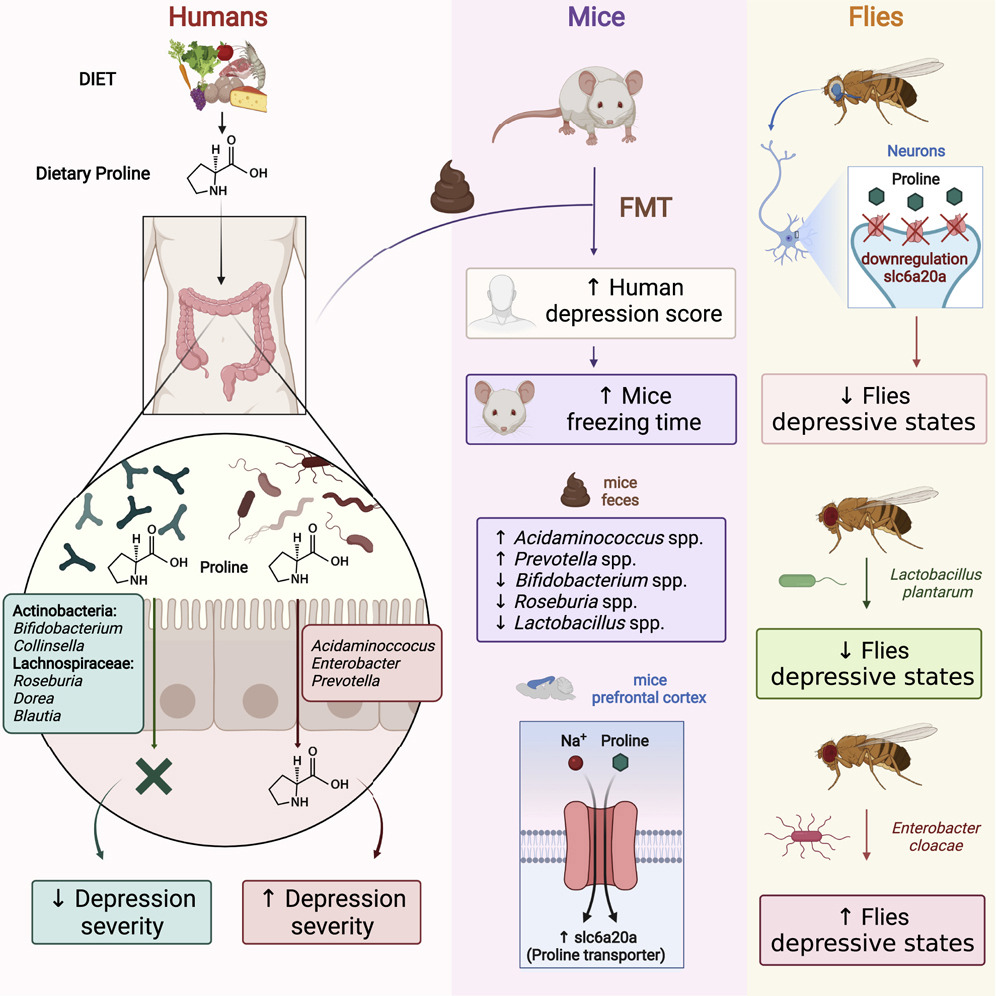
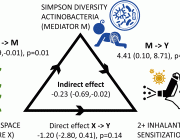
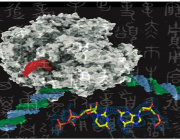
In this study we scanned 344 microbiomes from 1000- and 2000- year-old human teeth. The datasets originated from our previous studies on human ancient DNA (aDNA) and on microbial DNA accompanying human remains. We previously noticed that in many samples infection-related species have been identified, among them Tannerella forsythia, one of the most prevalent oral human pathogens. Samples containing sufficient amount of T. forsythia aDNA for a complete genome assembly were selected for thorough analyses. We confirmed that the T. forsythia-containing samples have higher amounts of the periodontitis-associated species than the control samples. Despites, other pathogens-derived aDNA was found in the tested samples it was too fragmented and damaged to allow any reasonable reconstruction of these bacteria genomes. The anthropological examination of ancient skulls from which the T. forsythia-containing samples were obtained revealed the pathogenic alveolar bone loss in tooth areas characteristic for advanced periodontitis. Finally, we analyzed the genetic material of ancient T. forsythia strains. As a result, we assembled four ancient T. forsythia genomes - one 2000- and three 1000- year-old. Their comparison with contemporary T. forsythia genomes revealed a lower genetic diversity within the four ancient strains than within contemporary strains. We also investigated the genes of T. forsythia virulence factors and found that several of them (KLIKK protease and bspA genes) differ significantly between ancient and modern bacteria.
Conclusions
In summary, we showed that NGS screening of the ancient human microbiome is a valid approach for the identification of disease-associated microbes. Following this protocol, we provided a new set of information on the emergence, evolution and virulence factors of T. forsythia, the member of the oral dysbiotic microbiome.
News source: www.bmcgenomics.biomedcentral.com
Article Reference: Philips, A., Stolarek, I., Handschuh, L. et al. Analysis of oral microbiome from fossil human remains revealed the significant differences in virulence factors of modern and ancient Tannerella forsythia. BMC Genomics 21, 402 (2020). https://doi.org/10.1186/s12864-020-06810-9
Targeting Microbiota 2021 Congress
October 20-22, 2021 - Paris, France & Online
www.microbiota-site.com